A study using bitBiome’s single cell sequencing for the analysis of the human resident bacteria and antibiotic resistance genes has been published in the academic journal Microbiome.
The paper titled “A single amplified genome catalog reveals the dynamics of mobilome and resistome in the human microbiome” has been published by a research team from bitBiome and Waseda University.
In this study, single-cell genome analysis of 30,000 oral and intestinal bacteria was performed on 51 Japanese subjects, including patients with cancer and inflammatory bowel disease, as well as healthy individuals, the largest number in the world.
As a result, the genomes and genes of hundreds of bacterial species that were overlooked by conventional methods were discovered. In addition, the presence of antibiotic resistance genes and their “carriers” was revealed on an individual bacterial level, making it possible to investigate the exchange of genes between bacteria in detail.
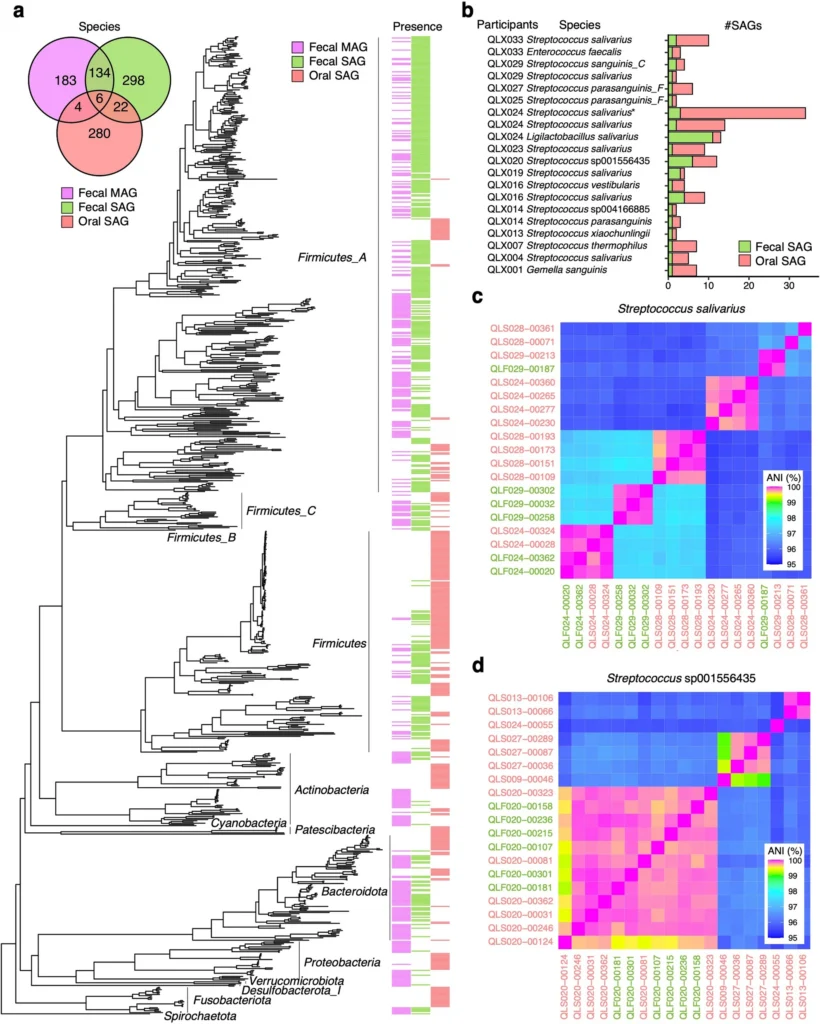
Microbial single-cell genome analysis, bit-MAP®, is expected to provide clues to a deeper understanding of how bacterial antibiotic resistance spreads and contributes to the development of personalized medicine targeting resident bacteria and new measures against antibiotic resistance.
For more details, please see the press release from Waseda University, “A new era of individual analysis of human resident bacteria: Decoding the genomes of 30,000 bacteria and tracking antibiotic resistance genes.”
Read the paper here: A single amplified genome catalog reveals the dynamics of mobilome and resistome in the human microbiome
■About bitBiome Inc.
bitBiome is a biotechnology company unlocking the full potential of our planet’s microbes to power the future of the bioeconomy. bitBiome’s platform is built on their proprietary single-cell microbial genome analysis technology, bit-MAP ®, which has enabled the creation of bit-GEM: an extensive and groundbreakingly diverse microbial database of over 2 billion sequences, sourced primarily from environmental samples and containing sequences not present in public databases. Leveraging their expertise in bioinformatics and machine learning, the company also offers a comprehensive enzyme discovery and engineering platform, bit-QED, which encompasses the identification, assessment, and modification of enzymes through wet lab evaluation and directed evolution. bitBiome is committed to improving existing biomanufacturing industries and creating new ones by delivering sequences and enzymes that cannot be found anywhere else. To learn more about bitBiome’s platform and services, visit bitbiome.bio.


